RESEARCH
Aims of our laboratory
The laboratory of cell biotechnology was established in April 2003 when the 2nd phase of the Bioproduction Engineering Research Center started. In our laboratory, we focus on various useful abilities of microorganisms, and aim to elucidate the universal principles underlying biological phenomena at the molecular level. We also aim to use these results for applied research to create more useful enzymes and compounds. Therefore, we analyze the function of enzyme and proteins, the regulatory mechanism of , using natural biochemistry, molecular biology, structural biology and protein engineering. The main research themes are introduced below.
Research on the mechanism and evolution of amino acid biosynthesis of microorganisms
Lysine is synthesized by the α-aminoadipate (AAA) pathway in yeasts and fungi via AAA as the biosynthetic intermediate, while is synthesized by the diaminopimelate (DAP) pathway via diaminopimelic acid in bacteria and plants. However, we discovered that the lysine biosynthesis of a thermophilic bacterium, Thermus thermophilus, synthesizes lysine by the AAA pathway.
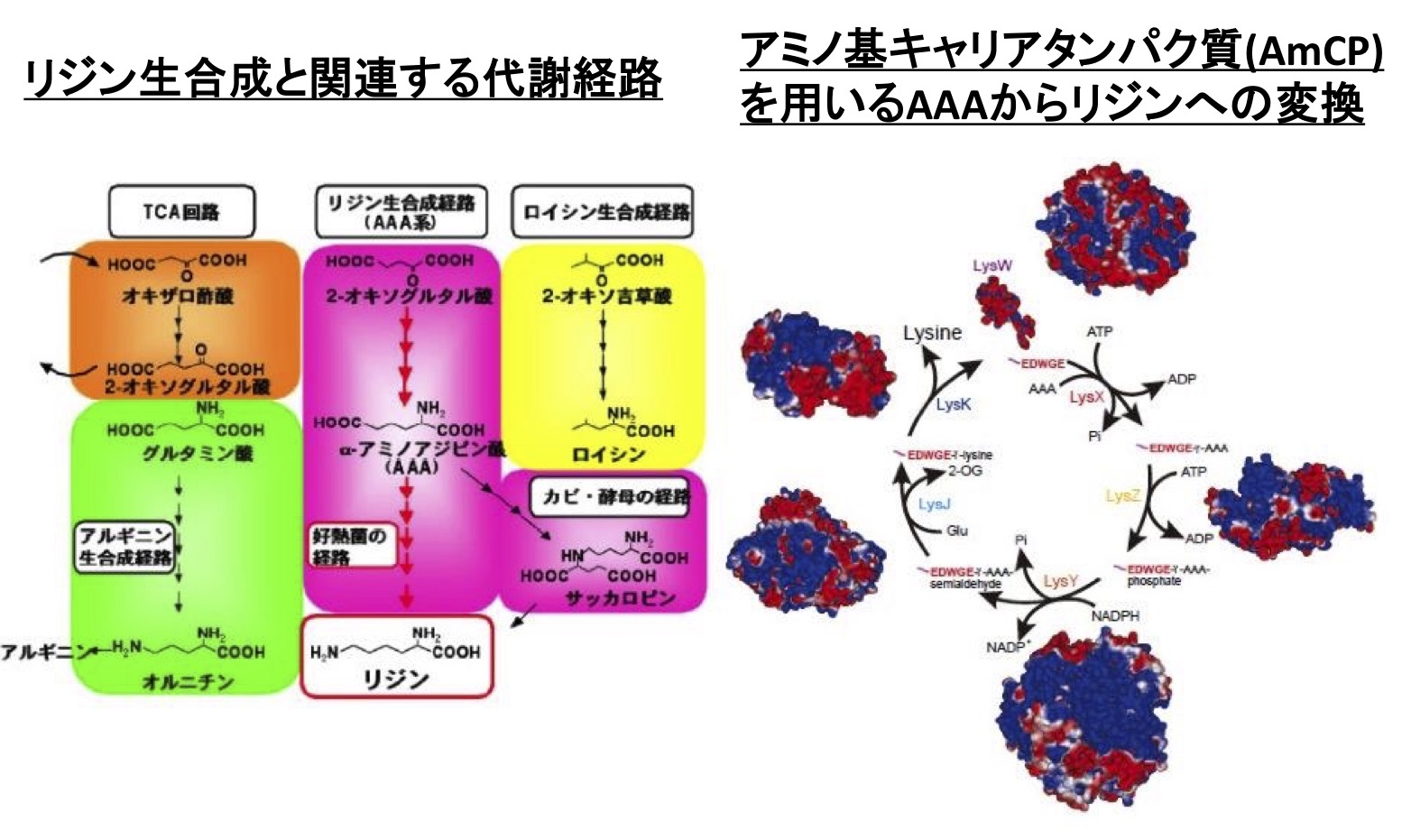
We also found that archaea which are considered to be closely related to the common ancestor of life have similar lysine biosynthesis pathways, and this biosynthetic system is not only for lysine but also for arginine. The enzymes involved in lysine biosynthesis have the characteristics that they have promiscuous substrate specificity. "Patchwork hypothesis" is one of the hypotheses of molecular evolution of enzymes. The hypothesis is that the existing repertoire of enzymes with strict substrate specificity is diverged from the single ancestral enzyme with promiscuous substrate specificity. According to this hypothesis, it can be considered that the present metabolic enzymes have been created by duplication, functional differentiation, and diversification of ancestral enzyme in the process of evolution. From this, it was thought that the lysine biosynthesis enzyme of the thermophilic bacterium had the remnant of the nature of the ancestral enzyme. We analyze the function and structure of such enzymes to elucidate the mechanism of substrate recognition of enzymes, and at the same time to clarify the molecular evolution mechanism of each enzyme and metabolism.
Exploration of diverse natural products biosynthesized via amino group carrier protein (AmCP)
In the course of study of the lysine biosynthetic pathway, we found that the latter half (conversion from α-aminoadipic acid to lysine) proceeds efficiently through protecting unstable biosynthetic intermediates using a carrier protein. This is the first discovery of carrier proteins involved in amino acid biosynthesis, and is also expected to be the basis for efficient amino acid fermentation production under high temperature conditions. Recently, we revealed that the similar system is also used for the production of secondary metabolites of actinomycetes and is involved in the production of novel non-proteinogenic amino acids.
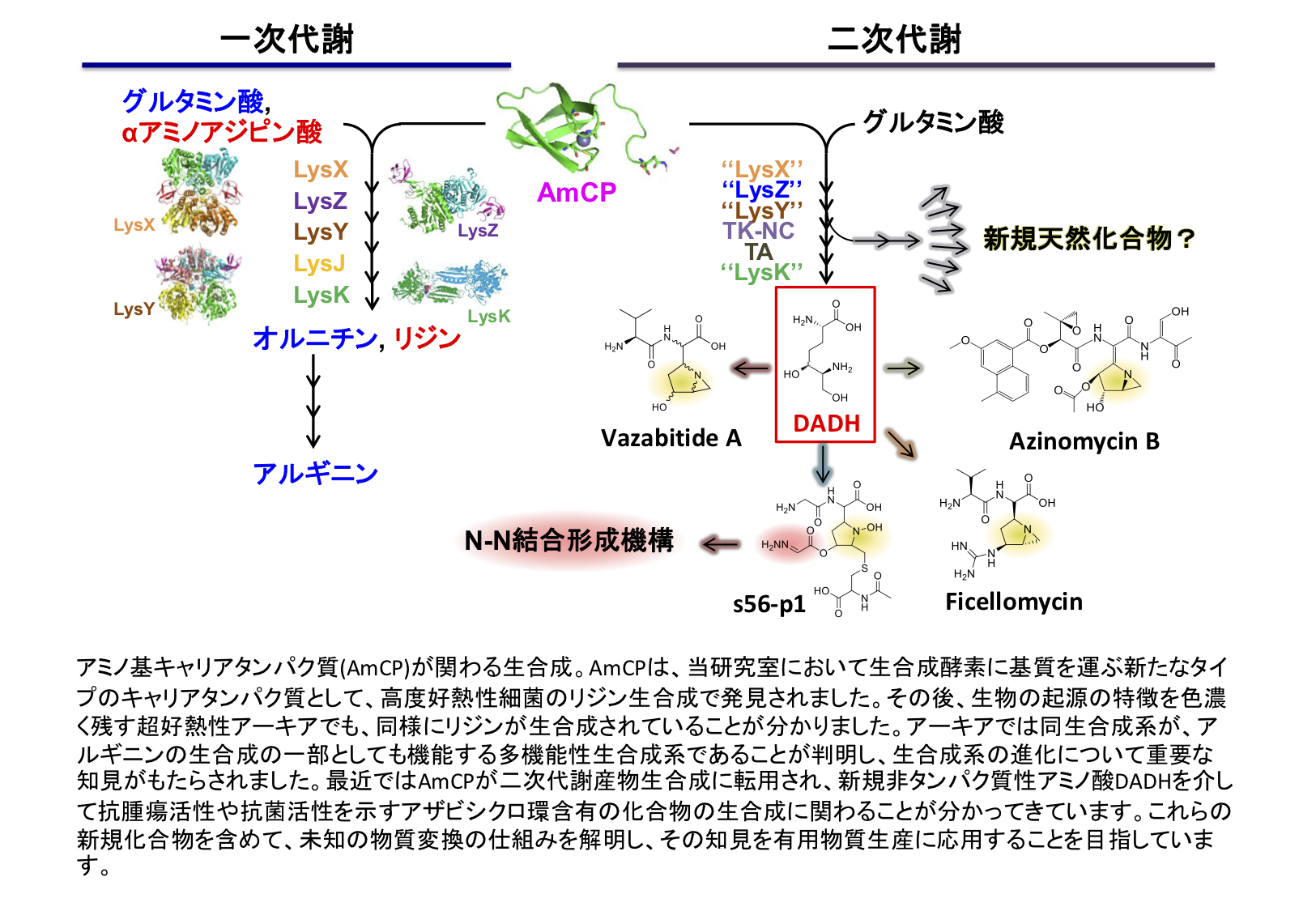
Database analysis suggests that similar systems exist in many other organisms, and we aim to elucidate their biosynthetic systems and produce new useful compounds. These researches are promoted as Grants-in-Aid for Scientific Research (Basic research (S)), “Elucidation of substance conversion mechanism using amino group-carrier protein and the application for production of useful compounds”.
Research on protein lysine acylation that regulates the protein and cellular function in bacteria
Protein lysine acylation, the most representative one is acetylation, is emerging as a ubiquitous post-translational modification found in organisms from bacteria to humans. Protein acetylation mostly targets the Nε-amino group of lysine residues enzymatically or non-enzymatically utilizing acyl-CoAs and acyl-phosphates as the acyl group donor. We found that lysine acetylation and succinylation (frequently occurring acyl-modifications) are dynamically changed in response to nutrient conditions and bacterial growth phases. We have revealed the effect of acetylation and succinylation on the functions of phosphoenolpyruvate carboxylase (PEPC), an inhibitor of 2-oxoglutarate dehydrogenase (OdhI), and elongation factor Tu. Our goal is to explore the impact and reveal the molecular mechanism of protein acylation on the cellular function and metabolism.

Study on control mechanism of enzyme function through protein interaction
Enzymes involved in amino acid biosynthesis have long been known to be feedback-regulated by the end products of biosynthesis. Recently, it became clear that cells also sense the environment and nutritional status of them.
We have determined the crystal structure of aspartate kinase from Corynebacterium glutamicum (CgAK), a key enzyme for industrial lysine fermentation, and clarified the molecular mechanism of the lysine high production.
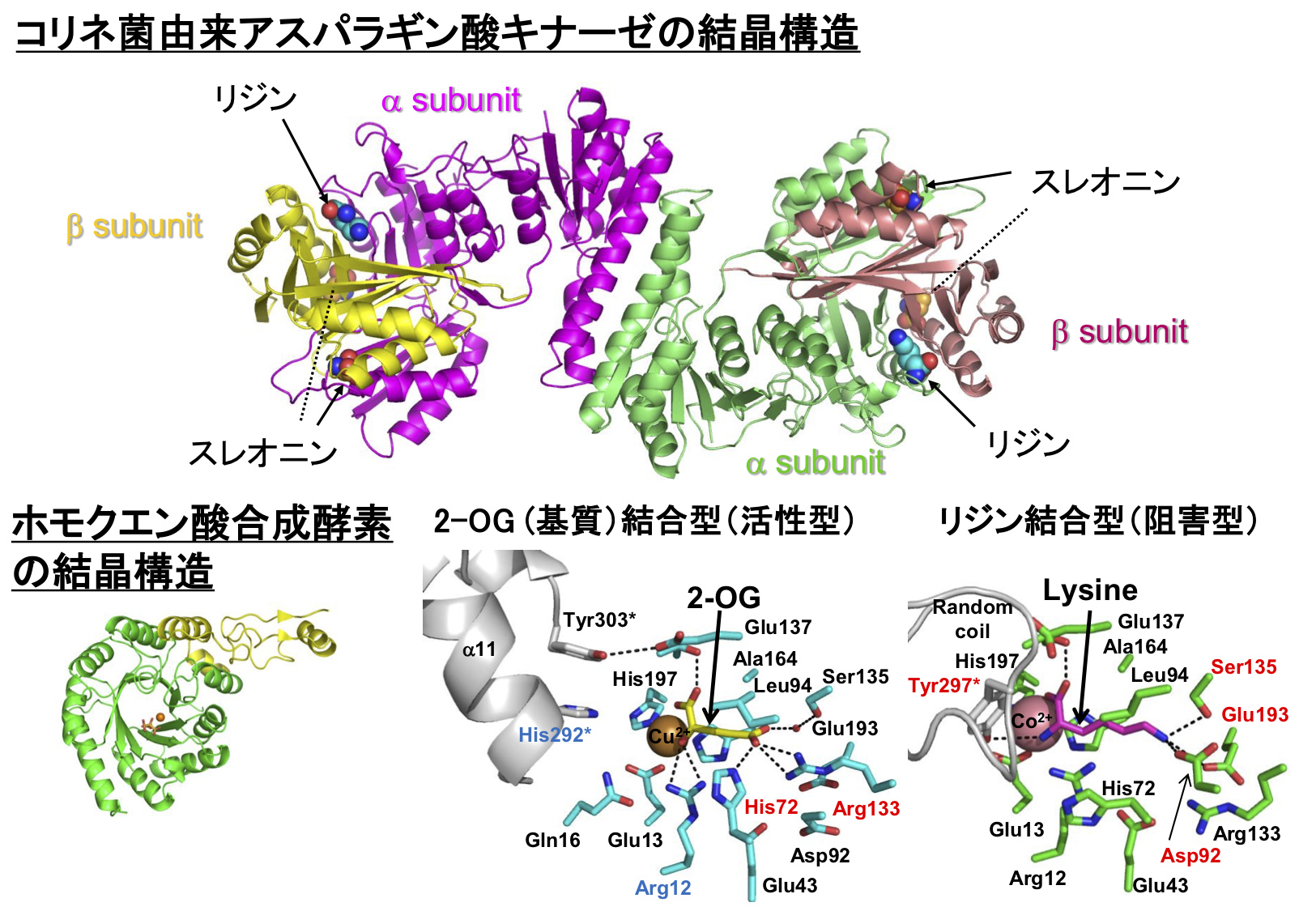
We also determined the crystal structure of homocitrate synthase (HCS), the first enzyme of lysine biosynthesis in thermophilic bacterium, Thermus thermophilus, and clarified mechanism of the unique feedback inhibition. We also aim the design to modification to highly activated and highly functional enzymes by conducting detailed analysis of the catalytic mechanism and regulatory mechanism using the three-dimensional structural information of the enzyme.
In addition, we focus on the interactions between metabolic enzymes and other proteins, and are exploring and analyzing new metabolic enzyme regulatory mechanisms. By clarifying such regulatory mechanisms, we believe it will be possible to control the metabolic flux of cells. Based on the findings obtained in this way, we are conducting research with a view to building a new substance production system.