





Algae are the most important contributors to the environment because they perform 40% of photosynthesis on earth. Algae are also the most diverse organisms with over 0.2 million species and varied morphologies.
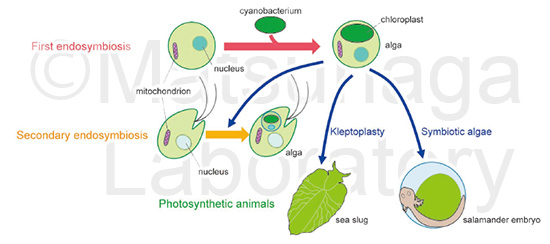
About 1 to 2 billion years ago, a primitive eukaryotic cell took up a cyanobacterium in the first endosymbiosis and evolved into algae with plastids derived from the endosymbiotic cyanobacterium. Several algae have become nested within secondary host cells through secondary endosymbiosis. Secondary endosymbiosis has occurred individually and horizontally at different times in different lineages in the animal, fungi and plant kingdoms, resulting in a great diversity of eukaryotes. Thus, secondary endosymbiosis is a powerful system by which heterotrophs acquire photosynthetic capability and becomes autotrophs.
Photosynthetic animals are found in different phyla, including Acoelomorpha, Chordata, Cnidaria, Mollusca and Porifera. Their heterotrophic lifestyles are partially supported by oxygen and energy supplied by photobionts, i.e., symbiotic algae and cyanobacteria. Symbiotic green algae in the cytoplasm of a salamander embryo supply oxygen and photosynthates for embryo development. Kleptoplasty is a unique symbiotic association in which sea slugs intracellularly retain functional chloroplasts extracted from predated algae. Evolutional insights from secondary endosymbiosis and extant photosynthetic animals imply that heterotrophic organisms have the potential to uptake algae and convert to autotrophic organisms.
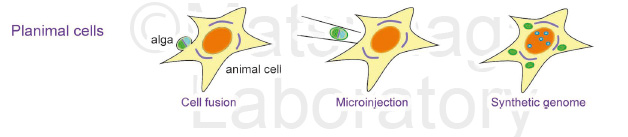
Professor Matsunaga proposed that artificial photosynthetic animal cells should be called “planimal cells” (2018). Strategies for the construction of planimal cells include three main approaches. One is to generate plant-animal hybrid cells by cell fusion techniques. The second approach is microinjection of algae or photosynthetic bacteria into animal cells. The third approach is the generation of chimeric chromosomes derived from the genomes of both algae and animals. We are challenging to create planimal cells based on the above three approaches.
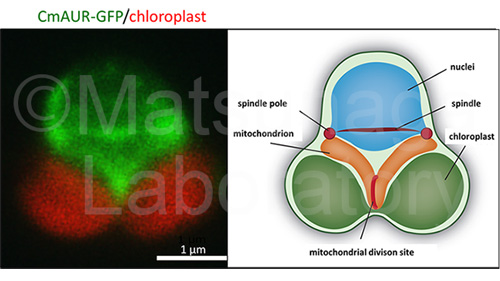
We use several microalgae for the construction of planimals cells. For example, a primitive alga, Cyandioschzon merolae is a candidate, which possesses a minimum set of organelles, sole nucleus, sole mitochondrion and sole chloroplast.
References
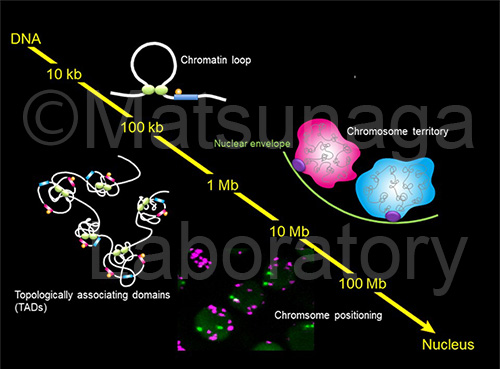
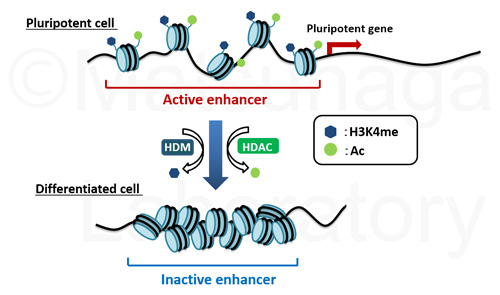
Several chromosome conformation capture techniques, which can quantify chromatin contacts, have disclosed the folded state of genomes in interphase nuclei. In particular, a chromosome conformation capture technique in combination with deep sequencing, Hi-C, can reveal the spatial genome organization in the whole genome through the frequency of the DNA interaction with each other. Chromosome conformation capture techniques also proposed a chromosomal module at sub-Mb scale, which is termed a topologically associated domain (TAD). TADs are more discrete chromatin structures consisting of chromosome territory.
We analyzed plant subnuclear dynamics with the following techniques.
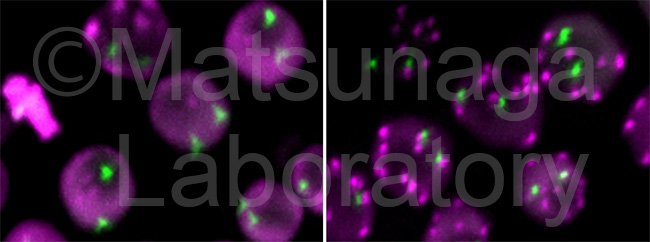
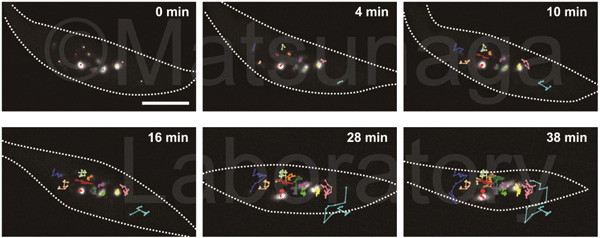
Among visualization techniques for chromatins in eukaryote nuclei, chromatin tagging systems contribute considerably to the analyses for chromatin dynamics. This system was constructed from bacterial operator/repressor systems using a repressor fused with a fluorescence protein. The tandem array of an operator sequence is introduced into the specific loci and the repressor with a fluorescent protein binds to the tandem array. Using this system, we identified RAD54 responsible for plant subnuclear dynamics.
References
Live imaging methods based on programmable DNA binding proteins derived from genome editing techniques were developed. These methods does not require the insertion of exogenous DNA sequences but depends on the interaction between cis elements (endogenous DNA sequences) and trans regulatory proteins (DNA binding proteins).
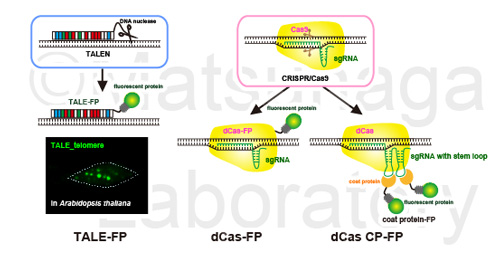
We successfully visualize telomeric sequences, 180-bp centromeric sequences, 28S and 5S rDNA sequences in Arabidopsis thaliana. We also succeeded in transient expression of dCas9-FP and single guide RNA (sgRNA) for telomeric sequences efficiently labeled telomere repeats in tobacco.
References
Hi-C (high-resolution chromosome conformation capture) provides the genome-wide interactions of all genomic regions. Hi-C can identify topologically associating domains and enhancer-promoter loops.
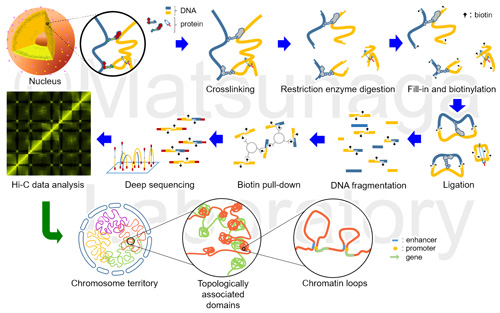
We performed Hi-C in plant nuclei and compared among several mutants in the response to environmental stresses.
References
Plant has a high regenerative capacity as is shown in tissue culture, where an entire organism can regenerate from a small piece of plant tissue under appropriate culture condition.
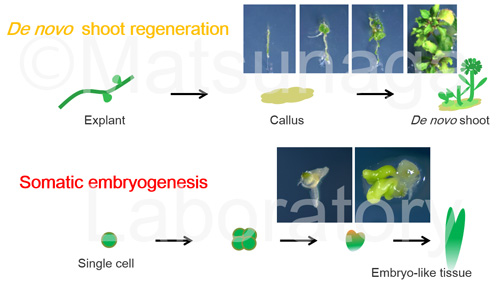
In the commonly used in vitro regeneration system of Arabidopsis thaliana, explants are first induced to form calli, followed by shoot induction. It has long been suggested that an acquisition of regenerative competency takes place during callus formation, but still only few reports described the molecular bases of this phenomenon. We are trying to analyze plant totipotency with epigenetic regulation.
Somatic embryogenesis is one of the best examples that demonstrate the high plasticity of plant. The formation of somatic embryos has been known as the evidence of totipotency of plant cells. We also characterized the key genes for somatic embryogenesis.
References
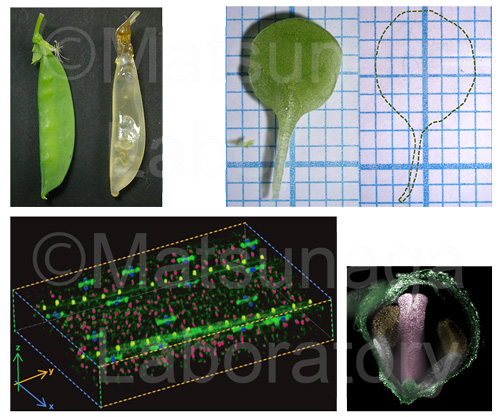
Clearing techniques eliminate factors that interfere with microscopy, including light scattering and absorption by pigments and cytoplasmic components. They allow fluorescence-based in-depth analyses of materials and characterization of the three-dimensional (3D) structures of organs. We developed a simple and rapid clearing and imaging method called “TOMEI” (Transparent plant Organ MEthod for Imaging), which enables the study of intact plant organs. You can commercially get TOMEI-kit from the following site.
http://www.tcichemicals.com/eshop/en/jp/commodity/T3530/ (English)
http://www.tcichemicals.com/eshop/ja/jp/commodity/T3530/ (Japanese)
References