理化学研究所 生命機能科学研究センター / 東京大学 生物普遍性研究機構
古澤研究室のウェブページへようこそ
生物システムは、様々な環境変化や内部状態の揺らぎの下で機能し続けられる頑強性(ロバストネス)を持つ一方で、 環境変化などに対して柔軟に内部状態を変化させる可塑性を持っています。このロバストネスと可塑性が両立できるという点は、 生物システムと人工システムの本質的な違いの一つです。しかし、どのようにして複雑な化学反応のネットワークがその両立を可能とするか、 メカニズムの理解は進んでいません。一方で、大規模な生物実験データが取得できるようになり、 そうしたデータに基づいてシステムの状態遷移やそのロバストネスを議論できるようになりつつあります。我々の研究チームでは、微生物の適応進化や、多細胞生物の発生過程などを題材として、細胞状態のロバストネスと可塑性について、 理論研究と実験研究の双方からの理解を目指しています。
細胞モデルを用いた
進化ダイナミクスの解析
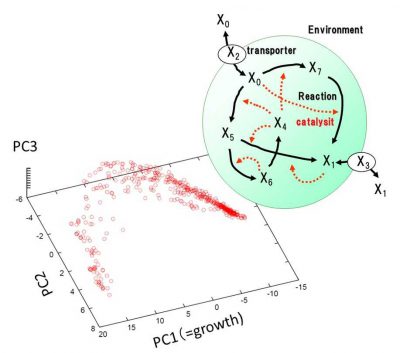
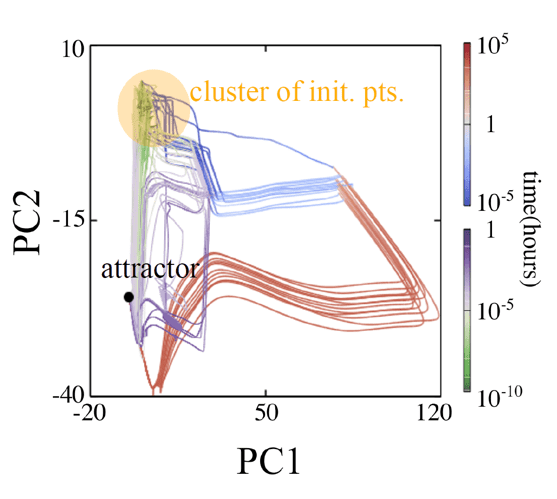
内部に簡単な代謝反応ネットワークを持つ細胞モデルの進化シミュレーションを行い、進化の結果として取り得る表現型が低次元に拘束されることを見出しました。
バクテリア遺伝子発現の
揺らぎと進化
大腸菌のそれぞれの遺伝子について、同一遺伝子型での発現量の揺らぎ、環境変動に対する応答、そして進化的応答を定量し、それら間にある関係性を解析しています。
代謝ダイナミクスの
理論的理解
微分方程式を用いて大腸菌の代謝反応システムをモデル化し、様々な環境下でどのような挙動を示すのかを力学系やネットワーク理論を援用しながら包括的に解析しています。
News & Information
- 金井さんが博士号を取得しました。4月から東京大学総合文化研究科に特任助教として赴任します。おめでとうございます!!修了おめでとうございます!
- 坂さん・小野さん・西田さんが修士課程を修了しました。おめでとうございます!修了おめでとうございます!
- 小野さんが日本生態学会大会ポスター賞(優秀賞)を受賞しました!
- 内田さんの発生過程の安定性を解析した論文がEvolution & Developmentに掲載されました。論文へのリンク
- 前田さんの抗生物質への耐性進化実験についてのレビューがAntibioticsに掲載されました。論文へのリンク
- 中村さんの幹細胞分化過程における階層性・不可逆性の出現メカニズムについての論文がPNAS Nexusに出版されました。論文へのリンク
- 佐藤匠哉さんの薬剤耐性予測についての論文がPhysical Review Researchに掲載されました。論文へのリンク
- 金井さんのバクテリアゲノム進化におけるトランスポゾンの役割についてのレビューがCurrent Opinion in Systems Biologyに掲載されました。論文へのリンク
- 堤さんが博士号を取得しました。10月から広島大学に特任研究員として赴任します。おめでとうございます!!
- 岩澤さんの大腸菌進化実験による適応度地形の推定に関するレビューが、日本生物物理学会誌に掲載されました。大腸菌の多次元表現型計測に基づく適応度地形の推定古澤 力,岩澤 諄一郎,前田 智也 学会誌へのリンク
- 堤さんの機械学習を用いた生物形態解析に関する論文がNPJ Systems Biology and Applications に出版されました。論文へのリンク
- 大町さん・斉藤さんの遺伝暗号表の適応度地形に関する論文がPLOS Comp. Biol. に出版されました。論文へのリンク
- 一井さん・浅野さん・宮内さんが修士課程を修了しました。おめでとうございます!修了おめでとうございます!!
- 内田さんのメダカの発生過程を用いて発現揺らぎと進化能の相関を解析した論文がEvodevoに出版されました。論文へのリンク
- 金井さんがゲノム微生物学会ポスター賞を受賞しました!金井雄樹, 津留三良, 古澤力, 「挿入配列活性によるオペロン構造形成の実証実験」 (学会サイトへのリンク)
- 岩澤さんのPLOS Biologyの論文について日経新聞に記事が出ました「薬剤耐性の進化を予測 理研、ロボで実験自動化」記事へのリンク
- 岩澤さんの大腸菌薬剤耐性進化における適応度地形の推定についての論文がPLOS Biologyに出版されました。論文へのリンク プレスリリース
- 芝井さん、津留さんの大腸菌の遺伝子発現量進化についての論文がG3に出版されました。論文へのリンク
- 自己複製システムを用いた、RNA分子の進化過程における構造揺らぎと進化可能性についの論文がRNAに出版されました。東大先進機構の市橋研との共同研究です。 論文へのリンク
- カメムシと大腸菌の共生進化実験の論文がNature Microbiol.に出版されました。産総研の深津・古賀グループとの共同研究です。 論文へのリンク プレスリリース
- 芝井さんの温度勾配環境下での進化実験を可能とする新規デバイスに関する論文が、SLAS Technologyに出版されました。論文へのリンク
- ERATO深津共生進化プロジェクトの全体会議が鶴岡で行われ、津留さん・芝井さん・佐藤駿さん・前田さん・金井さん・浅野さん・小野さん・西田さん・古澤さんが参加をしました。津留三良「遺伝子の発現量の進化しやすさは環境応答性から予測できるか?」 芝井厚「大腸菌のin vitro実験進化における自動進化制御系の開発」 佐藤駿平「カメムシ共生進化大腸菌の再現進化実験」 前田智也「マイクロバイオー […]
- 姫岡さんがコペンハーゲンで行われた国際会議"Major ideas in quantitative microbial physiology"で発表をしました。会議情報のリンク
- 古澤さんが国際ワークショップ""Cell, Energetics, and information: New perspectives on nonequilibrium systems"で招待講演を行いました。会議へのリンク
- 内田さん・堤さん・中村さんがナポリで行われたEuro EvoDevo2022で発表をしました。Yoshiyuki Nakamura, Yusuke Himeoka, Nen Saito, Chikara Furusawa, "Evolution of hierarchy and irreversibility i […]
- 姫岡さんの栄養飢餓と増殖のlag-timeの関係を解析した論文がmSphereに出版されました。論文へのリンク
- 内田さんの表現型揺らぎと形態進化に関する論文がBMC Biologyに出版されました。東京大学のプレスリリースへのリンク 論文へのリンク
- 金井さんが生命の起原および進化学会学術講演会で発表をしました。金井雄樹, 津留三良, 古澤力「オペロン構造を基本とする遺伝子発現制御機構の起源に関するIDE 仮説の実証実験」 https://sites.google.com/view/origintohoku46/%E3%83%9 […]
- 姫岡さん・中村さん・一井さん・新美さん(学部生)が物理学会で発表をしました。中村能之, 姫岡優介, 斉藤稔, 古澤力「細胞分化の階層性と不可逆性:状態空間におけるアトラクターの分布との対応」 新美倫太郎, 古澤力, 姫岡優介「代謝ネットワークの構造によるジェネラリスト・スペシャリスト戦略の出現」 […]
- 薬剤耐性大腸菌の電顕画像の機械学習による解析の論文がFront. Microbiol.に出版されました。大阪大学 産業科学研究所の西野研究室との共同研究の、薬剤耐性大腸菌の電子顕微鏡画像を機械学習に供し、耐性獲得と形態変化の関係を解析した論文が出版されました。 論文へのリンク https://www.frontiersin […]