研究内容
生命分子間の相互作用は、非共有結合の組み合わせによる特異的な多点結合により形成されている。本研究室ではこれらの相互作用への寄与を明らかにするために、蛋白質間相互作用全般、抗体-抗原、受容体-リガンド、蛋白質-金属イオン、多量体形成蛋白質など様々な蛋白質を題材とした解析を行っている。アミノ酸等を用いた溶媒制御によって蛋白質をハンドリングしながら、部位特異的変異導入を活用し、等温滴定型熱量測定(ITC)、表面プラズモン共鳴測定(SPR)、示差走査型熱量測定(DSC)、示差走査型蛍光測定(DSF)、マイクロスケール熱泳動(MST)等の物理化学的手法を用いた速度論的/熱力学的解析、円偏光二色性測定(CD)をはじめとした分光学的解析、さらには質量分析および結晶構造解析といった手法を駆使して、多角度的に生命分子間相互作用を精密解析し議論することにより、特異性・親和性創出機構の解明を試みている。得られた物理化学的パラメータは、計算・情報科学を駆使することにより分子のダイナミクスやアンサンブルの観点を取り入れることにより、in vitroとin silicoを融合して医療・材料応用を目指した蛋白質設計や低分子設計にチャレンジしている。これらの知見を踏まえ、高機能かつ優れた物性を持つ機能性分子の開発や特定の分子間相互作用を制御する分子種の探索を通じて創薬基盤の構築へと繋げるとともに、生命における“相互作用”の本質を理解することを目指している。
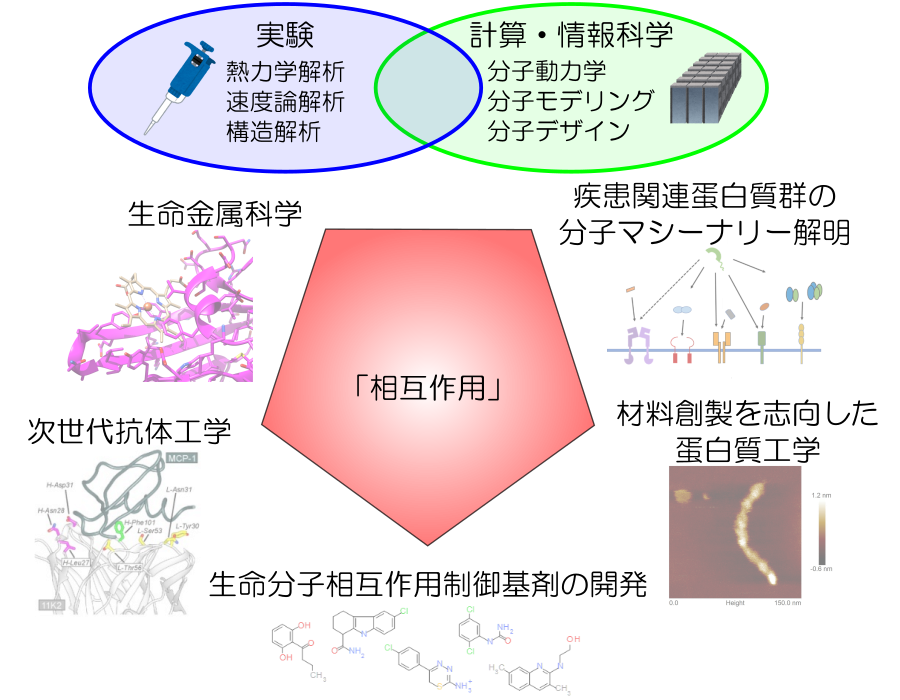
研究項目
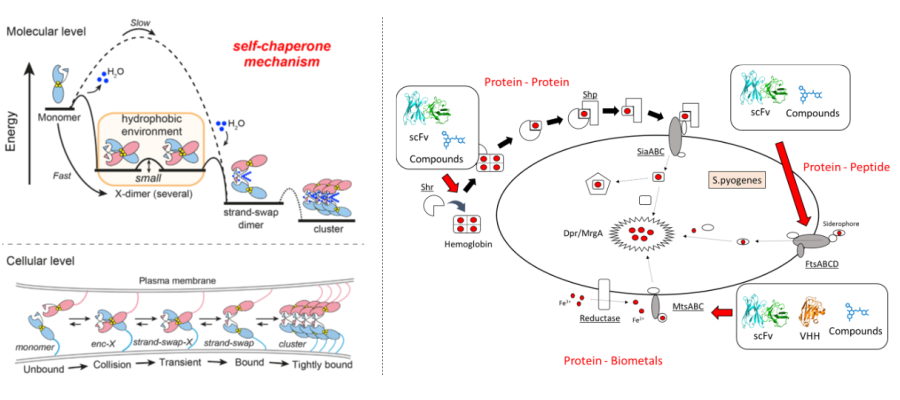
生命金属科学
金属は様々な生体分子の機能に深く根ざしています。ヒトをはじめとする動物から植物、微生物に至るまで多様な生物が有する蛋白質がその機能を発揮するために金属を必要とし、その金属依存的な機能が乱されることにより疾患へとつながります。例えば金属依存的に活性を発揮する蛋白質の制御が乱れることによって発がんやがんの転移につながり、その制御を正すことによってがんの治療へとつながると考えられます。また例えばヒトの体内で生育する病原性微生物が生存のために宿主であるヒトから金属を獲得するための精緻なシステムを作り上げており、そのシステムを理解、制御することにより、新規な作用機序に基づく抗菌剤の開発へとつながることが期待されます。すなわち、生体内における蛋白質機能と生命金属との関わりを明らかにすることは「生命」の理解、さらには様々な疾患の治療薬開発へとつながります。本研究室では金属依存的な細胞間接着や金属輸送関連蛋白質、病原性微生物の金属取り込み機構に着目し、金属を介した蛋白質機能の理解およびその制御を目指した研究を行っています。
研究テーマ例
- 金属依存的細胞接着蛋白質の会合体形成機構の解明
- 各種モダリティ分子を活用した金属依存的会合体形成の制御
- 病原性微生物の金属取り込み蛋白質群の機能解析と制御分子の開発
- 鉄輸送蛋白質認識抗体の分子認識機構の解明
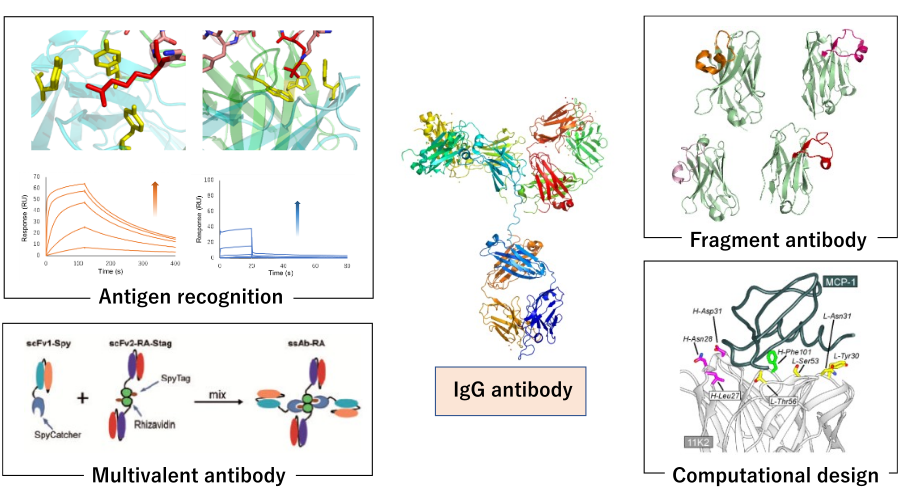
次世代抗体工学
抗体は抗原分子を高い親和性と特異性によって認識するため、バイオ医薬品や診断薬として多様な用途に使われています。今世紀に入り、抗体医薬を中心としたバイオ医薬が医薬品売上高上位を占めるようになり、一方で次世代の抗体には物性の改良、さらなる高機能化が求められるようになっています。本研究室では抗体の抗原認識を中心とした様々な物性について、多角度的な解析、エンジニアリングを行うことで、新時代に求められる抗体分子の創製に寄与する研究を行っています。
具体的には、抗体の分子認識機能、物性について、SPRやITCなどの物理化学的解析手法、構造生物学に加え、細胞生物学的手法を組み合わせ、抗原認識についてアミノ酸残基レベルで理解します。また、バクテリオファージの表面に分子を提示するファージディスプレイ法を活用しながらの進化工学による抗体の改変や、近年注目を集めている単ドメイン抗体の開発を行っています。さらに、抗体の小分子化、融合抗体の作製によるターゲティング能の変換、化学修飾による多機能化、計算科学を用いたデザインによる抗体の抗原親和性の向上を目指した研究も行っています。
研究テーマ例
- 抗体の分子認識メカニズムの解明:蛋白質抗原から低分子抗原まで(がん抗原蛋白質、オリゴペプチド、翻訳後修飾、金属錯体など)
- 合成ライブラリーを用いたヒト化単ドメイン抗体取得系の構築
- 抗体の抗原認識領域の構造に基づくペプチド設計法の開発
- 足場蛋白質を活用した部位特異的認識抗体の取得法構築
- 電荷改変による抗体表面デザイン
- 計算科学に基づいた合理的デザインによる抗体改良
- 低分子抗体の連結による多重特異性抗体の開発
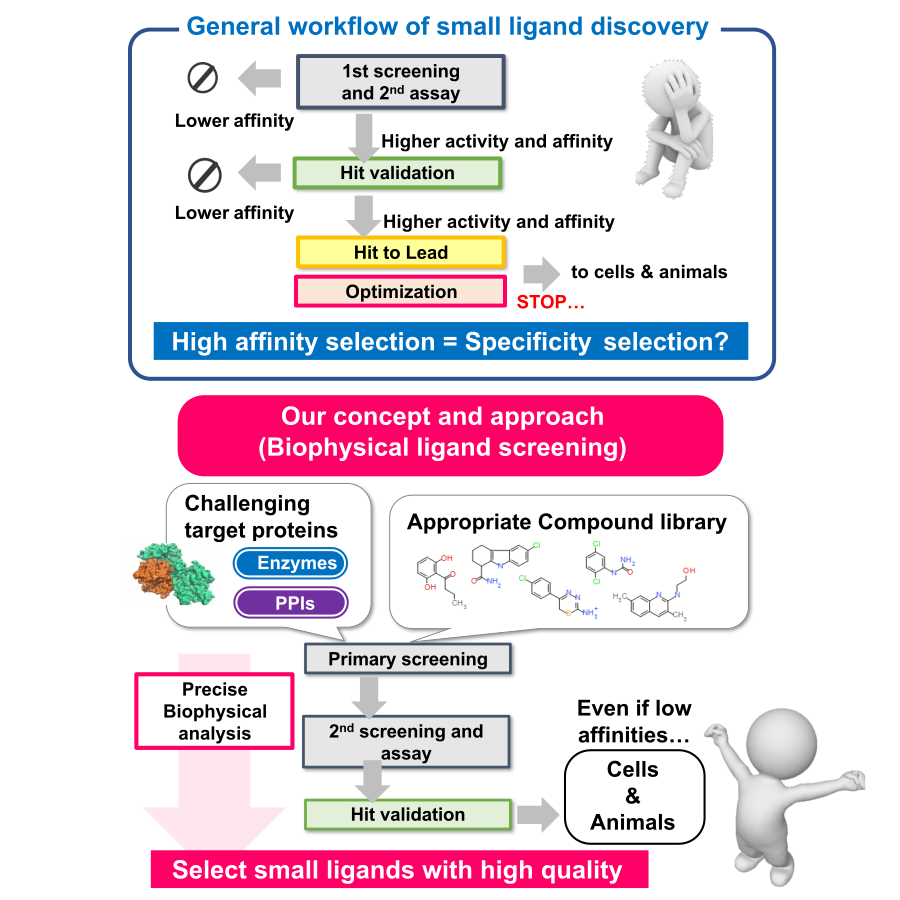
生命分子相互作用制御基剤の開発
生命分子相互作用を制御する低分子リガンドの探索とその最適化に関する研究を行っています。低分子薬剤の開発は、近年の標的分子の難易度の高さと多様化(膜蛋白質、蛋白質―蛋白質間相互作用、天然変性蛋白質など)により、標的分子の同定や調製、また探索技術(スクリーニング手法など)の構築といった開発初期の段階で滞る例が少なくありません。従来の探索技術では対応できないことも多いのが現状です。したがって生命分子の物性を正確に知り、その機能・相互作用を精密に解析できる手法を導入することで、新たな探索技術を開発・提案することが強く望まれます。そこで、探索(スクリーニング)、選抜(バリデーション)、そして最適化(ヒットtoリード、オプティマイゼーション)にて物理化学的解析手法(ITC, SPR, DSC, MST, HDX-MSなど)を活用し、結合親和性にとらわれることなく、相互作用の“質”を定量的に解析することによって、難易度の高い標的分子に対する新規な低分子リガンドの取得を試みています。フラグメントライブラリー、PPIライブラリー、天然物ライブラリーなど、さまざまなライブラリーを用いながら、本学創薬機構DDI、AMEDの創薬支援事業との緊密な連携のもと、治療薬等への応用のみならず、生命科学研究へ貢献する機能性低分子リガンドを創出します。
研究テーマ例
- 低分子リガンド探索における物理化学的解析技術の開発
- 蛋白質-蛋白質間相互作用PPI阻害剤の創出
- Fragment-based Drug Discovery
- 新規低分子薬剤に関する精密な相互作用解析
疾患関連蛋白質群の分子マシーナリー解明
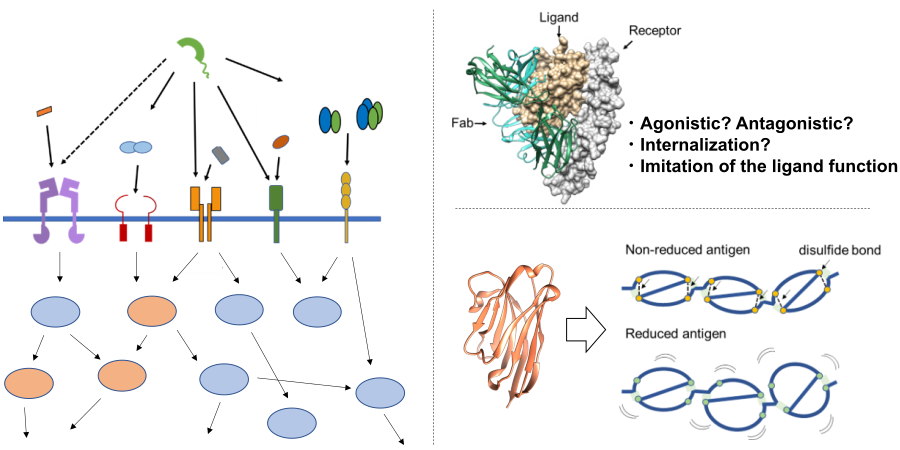
蛋白質は生体内において様々な生体分子と相互作用することにより生体システムを構築しており、そのシステムが破綻することにより、がんや自己免疫疾患等の重篤なものを含む様々な疾患を引き起こします。例えば、がん抑制蛋白質の発現が低下する、あるいは配列に変異が加わることにより発がんへとつながり、免疫関連蛋白質の過剰な活性化により自己免疫疾患を発症します。また、個々の生体内にとどまらず、感染症を引き起こす病原性微生物と宿主の間においても様々な蛋白質が相互作用しています。例えば近年重大な社会問題となっている新型コロナウイルスの感染においてもヒトの細胞表層に発現する受容体とウイルス表面の蛋白質との相互作用が重要な役割を果たしています。本研究室では、新型コロナウイルスを含む病原性微生物やがん関連蛋白質、自己免疫疾患に関連する蛋白質を中心に、疾患の原因となる各種蛋白質マシーナリーに関して、構造・機能解析を行っているほか、抗体や中分子、低分子等の各種モダリティ分子を活用した制御を目指した研究を展開しています。
研究テーマ例
- 新規がん抑制蛋白質によるがん抑制の分子機構解明と分子改変
- 抗体による受容体機能制御機構の解析
- 自己抗体の認識機構の解明を目指した還元・非還元抗原と抗体の相互作用解析
- 病原性微生物の表層蛋白質の機能解析と機能阻害分子探索
- がん関連蛋白質間の相互作用解析
材料創製を志向した蛋白質工学
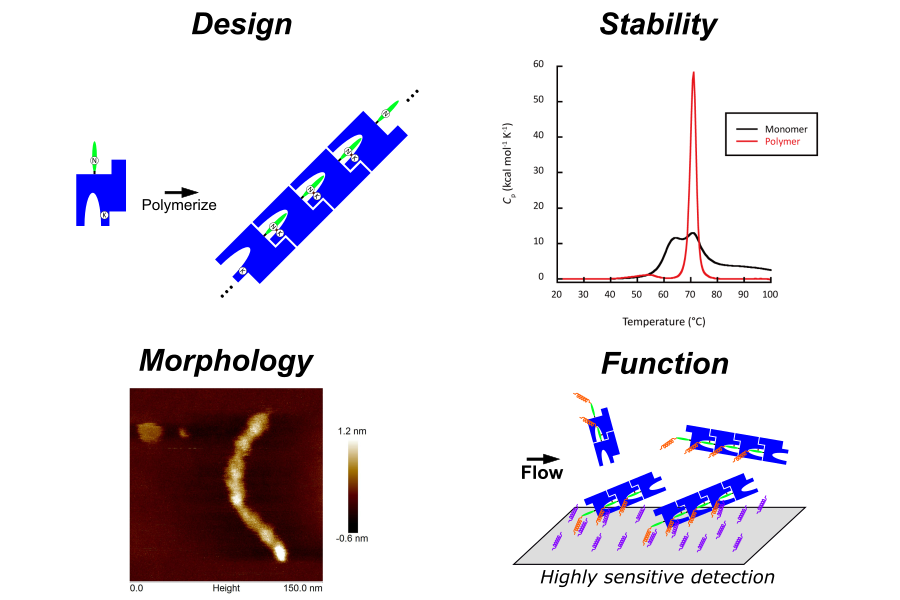
蛋白質を用いた機能性分子は医薬品のみならず、エネルギー産業や機能性材料など様々な分野への応用が期待されています。例えば毛髪は蛋白質の高度な自己組織化と線維化によって構成されており、また微生物の世界においても、蛋白質の重合によって形成された線毛と呼ばれるナノメートル径の超極細線維が菌体表層に存在します。これらの分子レベルでの蛋白質間相互作用解明によって、合理的な構造制御を可能とし、次世代の産業展開が期待されます。一方で、一般的に蛋白質は熱安定性およびコロイド安定性が低いために、汎用性材料への展開が困難となっています。そこで、本研究室では分子機械としての蛋白質高次構造を利用し、物性解析や計算化学を基により高機能かつ優れた物性を持つ分子種の合理的設計を目指した研究を行っています。
研究テーマ例
- 重合性蛋白質の設計に基づく機能性蛋白質高次構造体の創製と利用
- 分子内架橋による機能性蛋白質の安定化
- 標的に対して特異的共有結合を形成する蛋白質の分子認識メカニズム解明と応用
計算・情報科学に基づく生体分子の解析
上記の研究テーマに関連して、医科学研究所にあるスーパーコンピュータを用いて、計算・情報技術を用いた生体分子の解析を行っています。コンピュータを用いることで、分子の立体構造やその動的挙動を予測できます。また、生体分子に生じる変異の影響を定量的に議論することができます。コンピュータを用いることで、実験では実現不可能なスケールでの大規模アッセイも可能です。例えば、コンピュータを用いることで、200アミノ酸の蛋白質なら、その各残基を19種類のその他のアミノ酸に改変し、その影響を網羅的に調べることが、数時間で可能です。生体分子の解析以外にも、標的蛋白質に対する、医薬品候補化合物のバーチャルスクリーニングなども行っています。本研究室では、こうした計算結果に基づき実験を進めることで、より合理的に生命における“相互作用”の本質を理解することを目指しています。




