Research
Research topics
Interactions between biomolecules are formed by specific multipoint binding through a combination of non-covalent bonds. In order to clarify the contribution to these interactions, we are analyzing various proteins, including protein-protein interactions in general, antibody-antigen, receptor-ligand, protein-metal ion, and multimeric proteins. We are attempting to elucidate the mechanism of specificity and affinity creation by precisely analyzing and discussing biomolecular interactions from multiple angles using kinetic/thermodynamic analysis using physical chemistry methods such as isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), differential scanning calorimetry (DSC), differential scanning fluorescence (DSF), and microscale thermophoresis (MST), spectroscopic analysis including circular dichroism (CD), as well as mass spectrometry and crystal structure analysis. The obtained physicochemical parameters are used for the design of proteins and small molecules for medical and material applications by integrating in vitro and in silico approaches with computational and information science to incorporate the viewpoints of molecular dynamics and ensemble. Based on these findings, we aim to develop functional molecules with high functionality and excellent physical properties, and to explore molecular species that control specific molecular interactions, which will lead to the construction of a basis for drug discovery and the understanding of the nature of “interactions” in life.
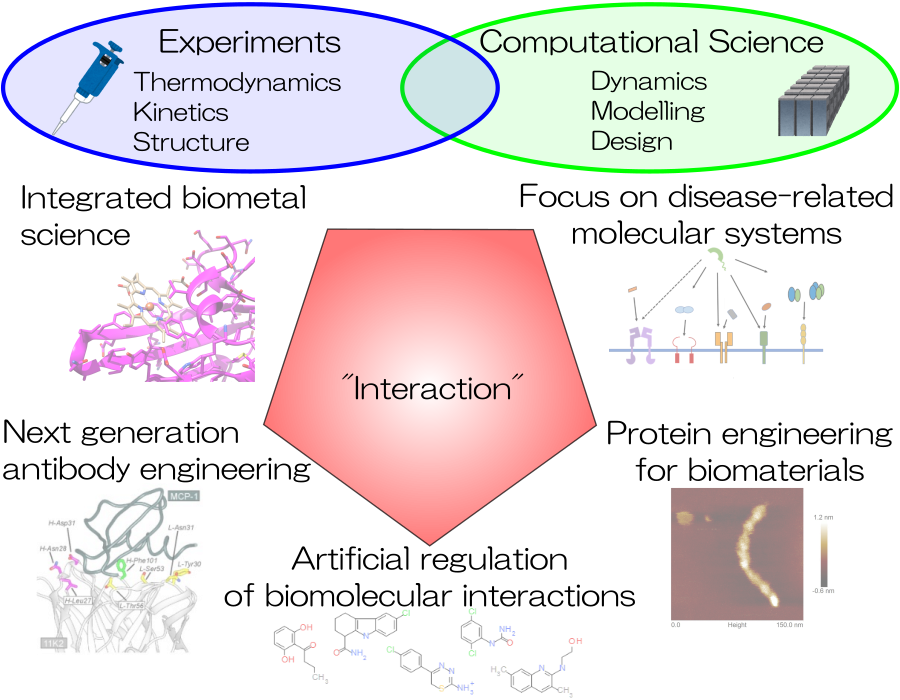
Research theme
- Integrated Biometal Science (IBmS)
- Next generation antibody engineering
- Artificial regulation of biomolecular interactions
- Focus on disease-related molecular systems
- Protein engineering for biomaterials
- Computational chemistry and biology for molecular design
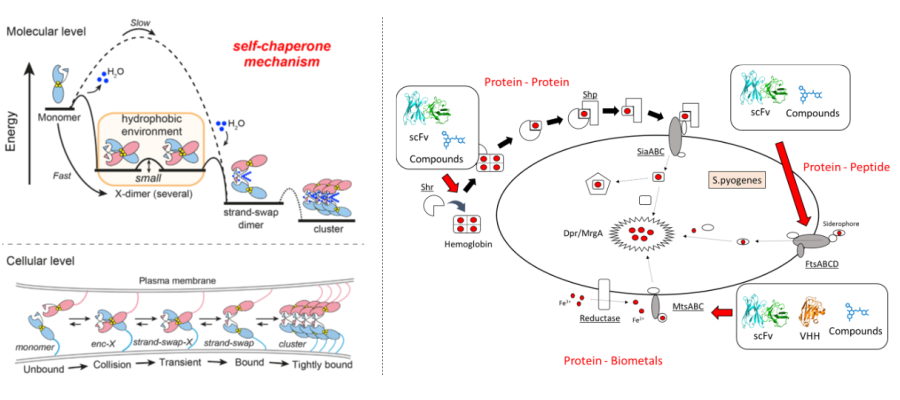
Integrated Biometal Science (IBmS):
Metals are deeply rooted in the functions of various biomolecules. Proteins from a wide variety of organisms require metals in order to function, and disruption of metal-dependent functions leads to diseases. For example, disruption of the regulation of proteins activated in a metal-dependent manner can lead to carcinogenesis and cancer metastasis, and correcting this regulation can lead to cancer treatment. For example, pathogenic microorganisms that grow in the human body have developed an elaborate system to acquire metals from their hosts in order to survive, and understanding and controlling this system is expected to lead to the development of antimicrobial agents based on novel mechanisms of action. In other words, clarifying the relationship between protein functions and metals in vivo will lead to a better understanding of “life” and the development of therapeutic drugs for various diseases. In our laboratory, we focus on metal-dependent cell-cell adhesion, metal-transport-related proteins, and metal uptake mechanisms of pathogenic microorganisms in order to understand and control metal-mediated protein functions.
Projects
- Elucidation of the mechanism of metal-dependent aggregate formation of cell adhesion proteins
- Regulation of metal-dependent aggregate formation by using various modality molecules
- Functional analysis of metal uptake proteins of pathogenic microorganisms and development of their regulatory molecules
- Elucidation of the molecular recognition mechanism of antibodies to iron transport proteins
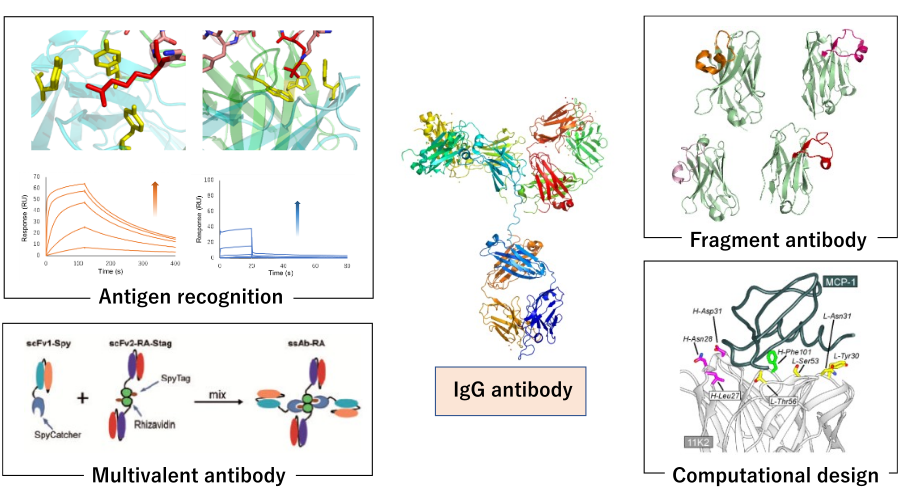
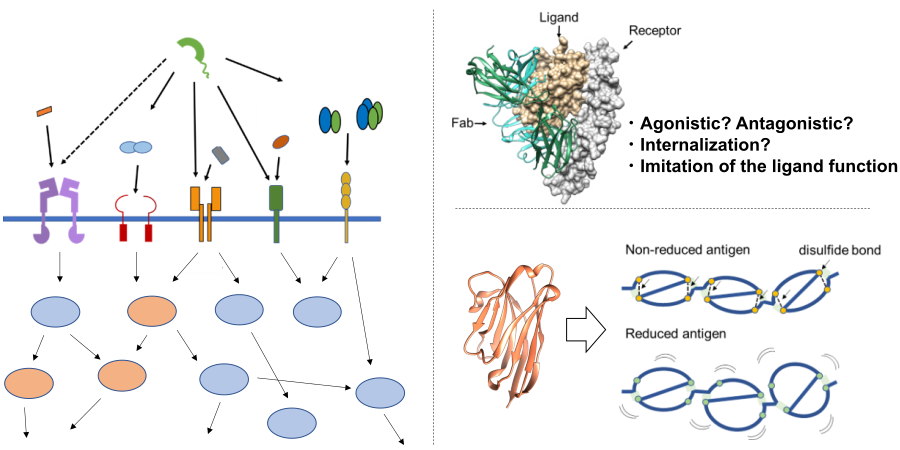
Next generation antibody engineering:
Since antibodies recognize antigens with high affinity and specificity, they are used in a variety of applications as biopharmaceuticals and diagnostic reagents. Since the beginning of this century, biopharmaceuticals, especially antibody drugs, have become one of the top sellers of pharmaceuticals, and next-generation antibodies are required to have improved physical properties and more advanced functions. In our laboratory, we are conducting research to contribute to the creation of antibody molecules required in the new era by analyzing and engineering various physical properties of antibodies with a focus on antigen recognition from various angles.
Specifically, we combine physicochemical analysis methods such as SPR and ITC, structural biology, and cell biology to understand the molecular recognition function and physical properties of antibodies at the amino acid residue level. In addition, we are modifying antibodies by evolutionary engineering using phage display method, which presents molecules on the surface of bacteriophages, and developing single-domain antibodies, which have attracted much attention in recent years. In addition, we are also working on small molecule antibodies, conversion of targeting ability by producing fusion antibodies, multifunctionalization by chemical modification, and improvement of affinity of antibodies by design using computational science.
Projects
- Elucidation of molecular recognition mechanisms of antibodies: from protein antigens to low molecular weight antigens (cancer antigen proteins, oligopeptides, post-translational modifications, metal complexes, etc.)
- Construction of a system to obtain humanized single-domain antibodies using a synthetic library
- Development of a peptide design method based on the structure of the antigen recognition region of antibodies
- Construction of a method for obtaining site-specific antibody recognition using scaffold proteins
- Antibody surface design by charge modification
- Improvement of antibodies by rational design based on computational science
- Development of multispecific antibodies by linking low-molecular-weight antibodies
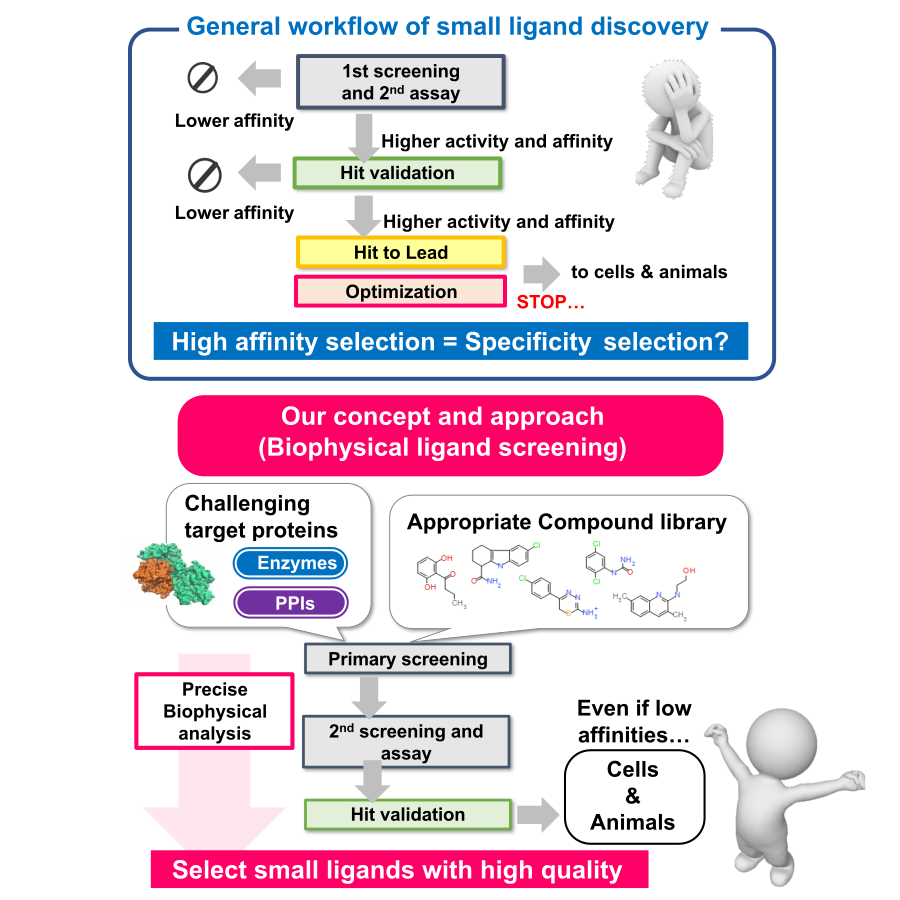
Artificial regulation of biomolecular interactions:
Our research focuses on the discovery and optimization of small molecule ligands that regulate biomolecular interactions. The development of small-molecule drugs often stalls in the early stages of development, such as the identification and preparation of target molecules and the establishment of discovery techniques (e.g., screening methods), due to the difficulty and diversification of target molecules (e.g., membrane proteins, protein-protein interactions, intrinsically disordered proteins) in recent years. In many cases, conventional search techniques cannot be applied. Therefore, it is highly desirable to develop and propose new search technologies by introducing methods that can accurately understand the physical properties of biomolecules and precisely analyze their functions and interactions. Therefore, physicochemical analysis methods (ITC, SPR, DSC, MST, HDX-MS, etc.) are utilized in screening, validation, and optimization (hit-to-lead, optimization) to quantitatively analyze the “quality” of interactions without being limited by binding affinity. Using a variety of libraries such as fragment libraries, PPI libraries, and natural product libraries, we will create functional small-molecule ligands that will contribute to life science research as well as therapeutic applications in close collaboration with Drug Discovery Initiative, The University of Tokyo and drug discovery support project of AMED.
Projects
- Development of physicochemical analysis techniques in small molecule ligand discovery
- Creation of PPI inhibitors for protein-protein interaction
- Fragment-based Drug Discovery
- Precise interaction analysis of novel small molecule drugs
Focus on disease-related molecular systems:
Proteins construct biological systems by interacting with various biomolecules in the body. When these systems fail, they can cause various diseases including serious ones such as cancer and autoimmune diseases. For example, decreased expression of tumor suppressor proteins or mutations in their sequences can lead to carcinogenesis, and excessive activation of immune-related proteins can cause autoimmune diseases. In addition, various proteins interact not only within individual organisms but also between pathogenic microorganisms that cause infectious diseases and their hosts. For example, the interaction between receptors expressed on the surface of human cells and proteins on the surface of the virus plays an important role in the infection of new coronaviruses, which have become a serious social problem in recent years. In our laboratory, we are conducting structural and functional analyses of various protein machineries that cause diseases, focusing on pathogenic microorganisms including new coronaviruses, cancer-related proteins, and proteins related to autoimmune diseases. In addition, we are developing research aimed at regulation using various modality molecules such as antibodies, medium molecules, and small molecules.
Projects
- Elucidation of molecular mechanism of cancer suppression by novel tumor suppressor proteins and molecular modification
- Analysis of the mechanism of receptor function regulation by antibodies
- Interaction analysis of antibodies with reducing and non-reducing antigens to elucidate the recognition mechanism of autoantibodies
- Functional analysis of surface proteins of pathogenic microorganisms and search for molecules that inhibit their functions
- Interaction analysis between cancer-related proteins
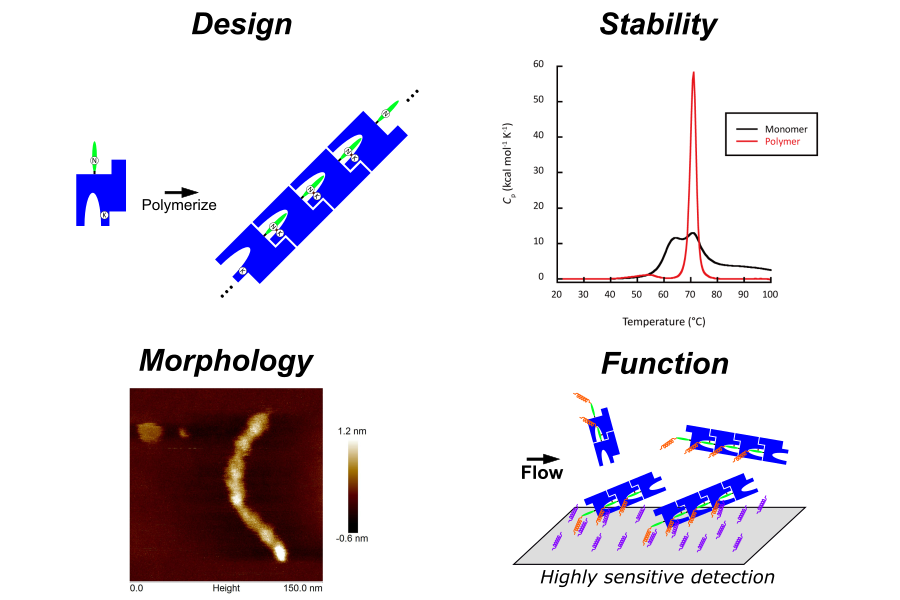
Protein engineering for biomaterials:
Functional molecules based on proteins are expected to be applied not only to pharmaceuticals but also to various fields such as the energy industry and functional materials. For example, human hair is composed of a high degree of protein self-assembly and fibrillation, and in the microbial world, ultra-fine fibers called pili, which are formed by the polymerization of proteins, exist in the surface layer of bacteria. Elucidation of these protein-protein interactions at the molecular level will enable rational structural control, which is expected to lead to next-generation industrial development. On the other hand, proteins generally have low thermal and colloidal stability, which makes it difficult to develop them into versatile materials. In our laboratory, we are conducting research on the rational design of molecular species with high functionality and excellent physical properties based on the analysis of physical properties and computational chemistry using the higher-order structure of proteins as molecular machines.
Projects
- Creation and utilization of higher-order structures of functional proteins based on the design of polymerizable proteins
- Stabilization of functional proteins by intramolecular cross-linking
- Elucidation and application of molecular recognition mechanisms for proteins that form specific covalent bonds to targets
Computational chemistry and biology for molecular design:
In relation to the above research themes, we are analyzing biomolecules using computational and information technology with the supercomputer at the Institute of Medical Science. By using the computer, we can predict the three-dimensional structure of molecules and their dynamic behavior. It is also possible to discuss quantitatively the effects of mutations on biomolecules. Computers can also be used to perform large-scale assays on a scale that cannot be achieved experimentally. For example, with the use of computers, it is possible to modify each residue of a 200 amino acid protein into 19 other amino acids and comprehensively study the effects of these modifications in a few hours. In addition to the analysis of biomolecules, we also conduct virtual screening of drug candidate compounds for target proteins. By conducting experiments based on these computational results, we aim to understand the nature of interactions in life in a more rational manner.




