ICON probe
The Best Way for Trapping Epigenetic Modification at the Specific Sequence
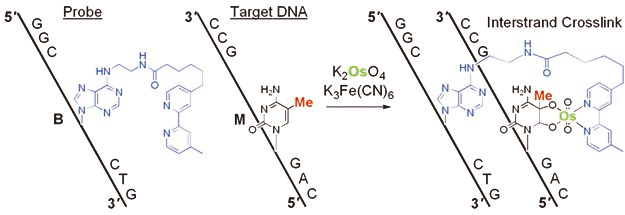
The development of a reaction for the detection of one epigenetic modification in a long DNA strand is a chemically and biologically challenging research subject. The formation of 5-methylcytosine (mC)–osmium complexes has been found as the basis for a bisulfite-free chemical assay for DNA methylation analysis. Osmium in an oxidized state reacts with C5-methylated pyrimidines in the presence of a bipyridine ligand to give a stable ternary complex. Sequence-specific osmium complex formation at mC was achieved by hybridization of a short DNA containing an adenine derivative with a tethered bipyridine moiety (ICON probe) to a target DNA sequence, and resulted in the formation of a crosslinked structure. This novel concept of methylation-specific reaction, based on a straightforward chemical process, expands the range of methods available for the analysis of epigenetic modifications. Advantages of the described method include amplification-insensitive detection, 5-hydroxymethylcytosine complexation, and visualization through methylation-specific in situ hybridization.
Principle
ICON probe contains an adenine derivative with a tethered bipyridine moiety, which forms a mismatched pair with mC. The mismatched pair is required for osmium-centered interstrand complex formation at the target mC. The formation of a mC/A mismatched pair causes partial disruption of π-stacking of the DNA duplex, and facilitates oxidation at the mC C5-C6 double bond, which is forced out of the duplex major groove. Combination of both the formation of a mismatched base pair together with an appropriately located bipyridine should result in complex formation at a specific mC, regardless of the presence of other reactive bases in a long DNA strand. ICON probes can form interstrand crosslinks through osmium complex formation, allowing sequence-selective mC labeling and obstruction of PCR amplification at the target mC residue.
Advantages
- – Sequence-selective
- – mC-positive
- – Simple detection process
- – No damage
- – Short time
- – Chemically labeled (fluorescence, biotin and more)
Targets of the Probes
5-Methycytosine (mC)
This is the major target of ICON probes. ICON technology is useful for identifying mC residues in target sequences at both the cellular and chromosomal levels, because ICON probes can label the target mC with fluorescent dyes, without causing serious DNA damage as observed with bisulfite methods and without the problems associated with nonsequence-specific mC binding as observed with anti-mC antibodies.
5-Hydroxymethylcytosine (hmC)
The hmC residue in DNA also showed high reactivity upon osmium-mediated oxidation. The reactivity of hmC in DNA was almost the same as that observed for 5mC in DNA. A stable hmC glycol-dioxidoosmium-bipyridine ternary complex was formed. Combination of the osmium-mediated oxidation and the hmC detection technologies with different chemical approaches will extend the range of applications in epigenetics research.
Thymine (T) -It is a special case.
– When the bipyridine-modified adenine locates at the terminus of the probe,
In the ICON probe, T does not form any osmium complex even when located opposite the bipyridine-modified adenine. However, when the location of a bipyridine-modified adenine within the ICON probe was varied, the reactivity and base selectivity of the osmium crosslinking changed. Osmium-mediated oxidation using the ICON probe containing a bipyridine-modified adenine at the end of the strand showed high crosslink yields for not only T opposite the bipyridine-modified adenine, but also T located at the unhybridized region of the target DNA.
The reaction of a bipyridine-end-labeled ICON probe can be used as a control in conjunction with a bipyridine-internal-labeled probe. For example, an ICON probe containing a Cy5 label at the 5’-end and a bipyridine-modified adenine inside the sequence (discrimination ICON), and an ICON probe containing a Cy3 label near the 3’-end and a bipyridine-modified adenine at the 5’-end (control ICON), which could recognize the 3’-neighbor of the sequence to which the discrimination ICON binds, were used to investigate test HoxA5 fragments that had been attached to beads. A combination of mC-specific crosslinking from bipyridine-internal ICON probes and nonspecific crosslinking from bipyridine-terminal ICON probes was effective for fluorometric detection of mC.
– When TmCG is the target sequence,
In the reaction with ICON probes, T is usually inactive against osmate because the bipyridine-modified adenine and T can form a Watson–Crick-like base pair in the duplex structure. However, osmium complex formation using the ICON probe results in oxidation of the T in the TmCG/CBA duplex (B: a bipyridine-modified adenine). Interestingly, whereas oxidation was observed at both T and mC for a TmCG sequence, oxidation was negligible at both T and C in a TCG sequence. The reason behind this difference in reactivity is not obvious. Clearly, the T base flanking a B/mC mismatched base pair may be more reactive than T in the normally stacking duplex because it is easier for the oxidant to access the T in the mismatched pair. As a result, although complex formation at the T of TmCG occurs, the function of the ICON probes, which discriminate mC from C using complexation efficiency, is maintained completely.
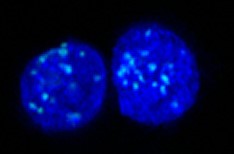
MeFISH (methylation-specific fluorescent in situ hybridization)
ICON technology is useful for identifying mC residues in target sequences at both the cellular and chromosomal levels. Professor Hiroyuki Sasaki and his group in Kyushu University applied ICON technology to develop a novel method, named methylation-specific fluorescence in situ hybridization (MeFISH), for visualizing the DNA methylation status at specific sequences in individual nuclei or chromosomes. The overall MeFISH protocol involves conventional FISH technology for sequence-specific target detection, and ICON technology for mC detection. Tissue sections, cells, or chromosome spreads are first subjected to FISH with the fluorescence-labeled ICON probes, and the location of fluorescent signals is then recorded. Next, the specimens are treated with osmium to induce mC-dependent crosslinking, and noncrosslinked probes are removed in a denaturation step. Subsequent comparison of FISH and MeFISH images allows an estimate of the methylation status.
The utility of MeFISH technology has been extended to the imaging of intact early embryos or small embryonic tissues by Professor Kuniya Abe and his group in RIKEN. The whole-mount MeFISH approach enables the observation of the DNA methylation status of satellite repeats in developing mouse primordial germ cells. Combination of whole-mount MeFISH with immunostaining or RNA-FISH facilitates the simultaneous visualization of DNA methylation and protein or RNA expression at single-cell resolution without destroying embryonic and nuclear structures.
Availability
ICON probe is commercially available.
GeneDesign Co. Ltd. (Ajinomoto Bio-Pharma Services). You can find “ICON probe” in the web page: https://www.ajioligos.com/en/products/nucleic-acid-medicine-oligos/
Papers to be referred
- Org. Biomol. Chem. 2006, 4, 1638-1640. (Os oxidation of mC (no ICON))
- Bioorg. Med. Chem. 2007, 15, 1615-1621. (Fluorescence quenching by Os Complex (no ICON))
- Bioorg. Med. Chem. Lett. 2007, 17, 1912-1915. (Degradation of DNA by bisulfite treatment (no ICON))
- J. Am. Chem. Soc. 2007, 129, 5612-5620. (mC labeling (no ICON))
- J. Am. Chem. Soc. 2007, 129, 14511-14517. (First paper on ICON)
- Org. Biomol. Chem. 2008, 6, 269-271. (mC-Os complex structure (no ICON))
- Coll. Symp. Ser. 2008, 10, 146-149. (Synthesis)
- Org. Biomol. Chem. 2008, 6, 3905-3907. (Reactivity of T)
- Org. Biomol. Chem. 2009, 7, 21-26. (Review)
- Bioconjugate Chem. 2009, 20, 603-607. (Reactivity of T)
- J. Synth. Org. Chem. Jpn. 2009, 67, 680-686. (Review (in Japanese))
- Org. Biomol. Chem. 2011, 9, 4176-4181. (hmC)
- J. Nucleic Acids 2011, 2011, 480570. (On-chip analysis)
- Curr. Protoc. Nucleic Acid Chem. 2011, 8.7.1-8.7.17. (Protocol)
- Chem. Biodiv. 2012, 9, 2000-2007. (Bipyridine incorporation site)
- Nucleic Acids Res. 2013, 41, e186. (MeFISH)
- PLoS ONE 2014, 9, e95750. (Whole-mount MeFISH)
- ChemMedChem 2014, 9, 1958-1965. (Review)
- ChemBioChem 2018, 19, 1653-1656. (Epitranscriptome (no ICON))
- Chem. Rec. 2018, 18, 1727-1744. (Review)